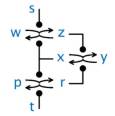
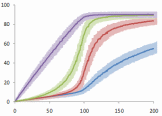
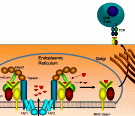
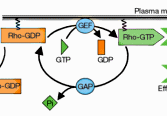
An abstract machine is a
fictional information-processing device that can, in
principle, have a number of different physical
realizations (mechanical, electronic, biological, or
software). Biochemical toolkits in cellular biology
(nucleotides, amino acids, and phospholipids) can be
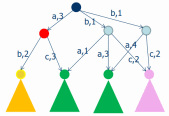
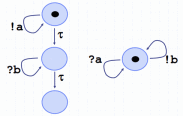
seen as abstract machines with appropriate sets of
states and operations, corresponding respectively to
genes, proteins, and membranes. To understand the
functioning of a cell, one must understand (at least)
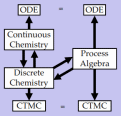
how the various machines interact. This involves
considerable difficulties in modeling and simulations
because of the drastic differences in the "programming
model" of each machine, and in the time and size scales
involved.